Tools
1. iBEAT V2.0 Cloud
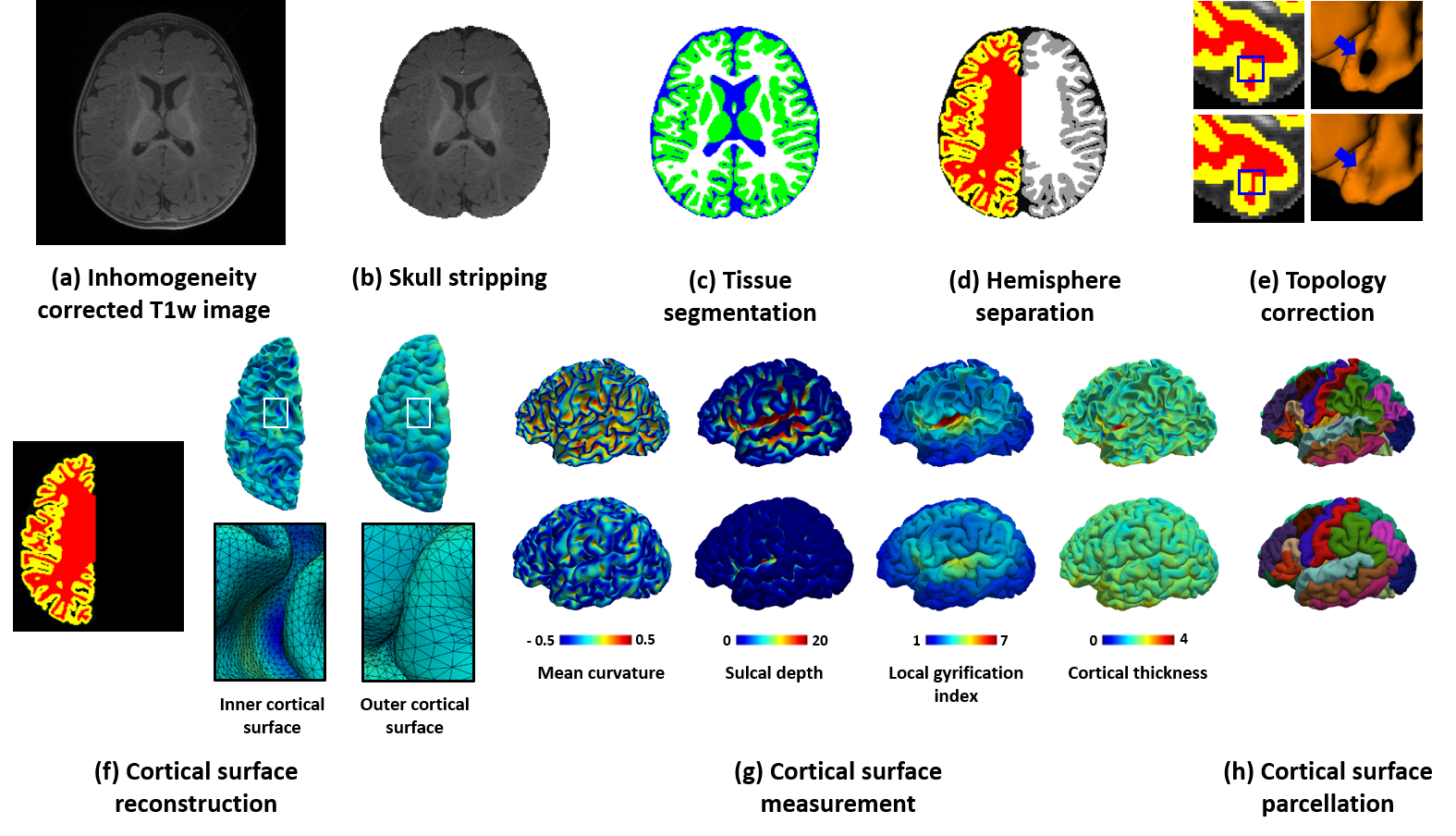 |
A new version of iBEAT (Infant Brain Extraction and Analysis Toolbox) is now available online as iBEAT V2.0 Cloud (http://www.ibeat.cloud/), which is developed by Dr. Gang Li’s and Dr. Li Wang’s teams with cutting-edge techniques (including deep learning). iBEAT V2.0 Cloud can handle pediatric brain images from different sites with various scanners and protocols. Users can process brain structural images from birth through adolescence, including images during the first postnatal year, which typically exhibit low tissue contrast and dynamic appearance and size changes. All uploaded data will be securely managed in the iBEAT V2.0 web server and will not be distributed to public. The current functionality of iBEAT V2.0 Cloud includes: preprocessing, inhomogeneity correction, skull stripping, tissue segmentation, left/right hemisphere separation, topology correction, cortical surface reconstruction, cortical surface measurement, and cortical surface parcellation. iBEAT V2.0 has successfully processed 12,000+ infant brain images from 90+ institutes and consistently achieves superior results. (Link) |
2. UNC 4D Infant Cortical Surface Atlases
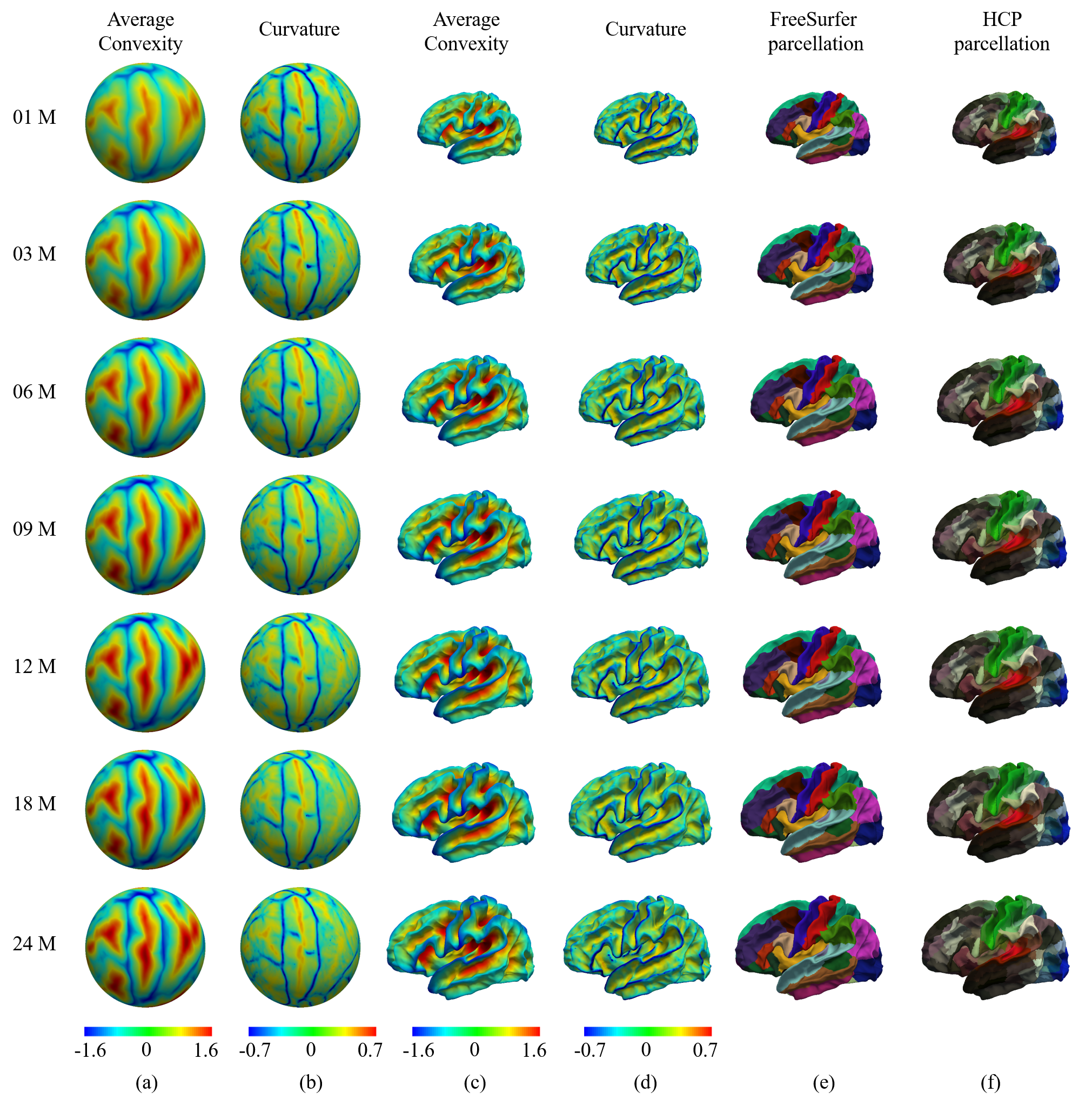 |
UNC 4D longitudinal infant surface atlases of cortical structures from neonates to 6 years of age contain 11 time points, including 1 month, 3 months, 6 months, 9 months, 12 months, 18 months, 24 months, 36 months, 48 months, 60 months and 72 months, thus densely covering and well characterizing the critical stages of the dynamic early brain development. (4000+ Downloads on NITRC) Construction of 4D High-definition Cortical Surface Atlases of Infants: Methods and Applications. Gang Li, Li Wang, Feng Shi, John H. Gilmore, Weili Lin, Dinggang Shen. Medical image analysis, vol. 25 (1), pp. 22-36, 2015. Learning 4D Infant Cortical Surface Atlas with Unsupervised Spherical Networks. Fenqiang Zhao, Zhengwang Wu, Li Wang, Weili Lin, Shunren Xia, Gang Li. International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2021, Strasbourg, France, Sep 2021. |
3. UNC Neonate Cortical Surface Atlases
 |
The UNC spatiotemporal neonatal cortical surface atlas focuses on newborn babies. Due to their rapid development, the atlas is built at each week, including surface atlases at 39, 40, 41, 42, 43, and 44 post-menstrual weeks. (Download on NITRC) Construction of Spatiotemporal Neonatal Cortical Surface Atlas using a Large-scale Dataset. Zhengwang Wu, Gang Li, Li Wang, Weili Lin, John H Gilmore, Dinggang Shen. IEEE International Symposium on Biomedical Imaging (ISBI), Washington, DC, USA, April 4-7, 2018. Construction of 4D Neonatal Cortical Surface Atlases Using Wasserstein Distance. Zengsi Chen, Zhengwang Wu, Liang Sun, Fan Wang, Li Wang, Fenqiang Zhao, Weili Lin, John H. Gilmore, Dinggang Shen, Gang Li. IEEE International Symposium on Biomedical Imaging (ISBI), Venice, Italy, Apr. 8-11, 2019. |
4. Spherical U-Net Package
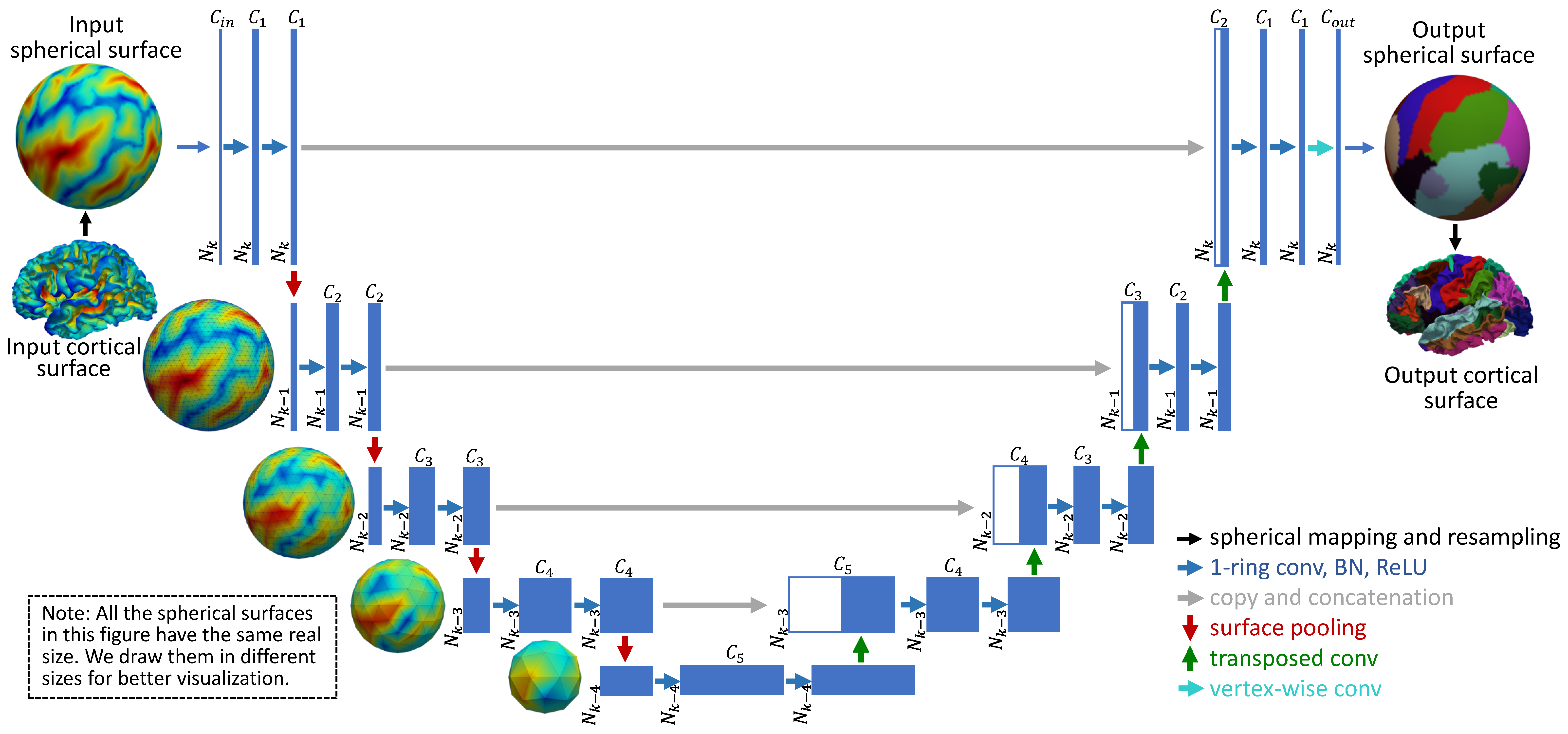 |
Python-based spherical cortical surface processing tools, including spherical resampling, interpolation, parcellation, registration, atlas construction, etc. It provides fast and accurate cortical surface-based data analysis using deep learning techniques. (Download on NITRC and GitHub) Spherical U-Net on Cortical Surfaces: Methods and Applications. Fenqiang Zhao, Shunren Xia, Zhengwang Wu, Dingna Duan, Li Wang, Weili Lin, John Gilmore, Dinggang Shen, Gang Li. Information Processing in Medical Imaging (IPMI), 2019. Spherical Deformable U-Net: Application to Cortical Surface Parcellation and Development Prediction. Fenqiang Zhao, Zhengwang Wu, Li Wang, Weili Lin, John H. Gilmore, Shunren Xia, Dinggang Shen, Gang Li. IEEE Transactions on Medical Imaging (TMI), 2021. |
5. UNC-LPBR 4D Cynomolgus Macaque Atlases
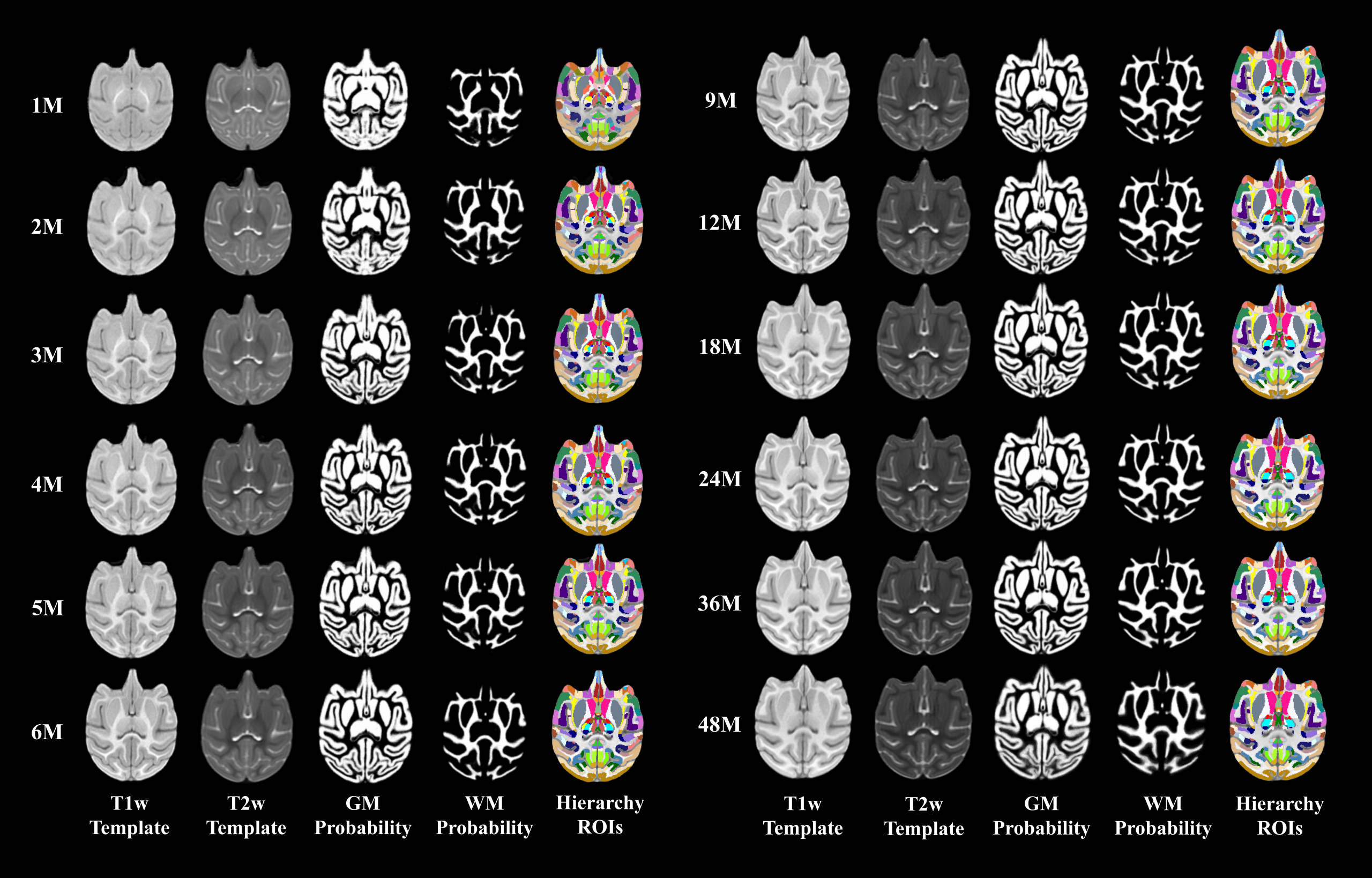 |
To study the early dynamic postnatal brain development of cynomolgus macaques, we construct the first set of spatial-temporal (4D) brain atlases and associated tissue probability maps with totally 12 time-points (i.e., 1, 2, 3, 4, 5, 6, 9, 12, 18, 24, 36, and 48 months of age) based on 175 longitudinal structural MRI scans from 39 typically-developing cynomolgus macaques. Meanwhile, to facilitate region-based analysis using our atlases, we also provide two popular hierarchy parcellations, i.e., cortical hierarchy maps (6 levels) and subcortical hierarchy maps (6 levels) on our 4D macaque brain atlases. (Download on NITRC) Longitudinal Brain Atlases of Early Developing Cynomolgus Macaques from Birth to 48 Months of Age. Tao Zhong, Jingkuan Wei, Kunhua Wu, Liangjun Chen, FenqiangZhao, Yuchen Pei, Ya Wang, Hongjiang Zhang, Zhengwang Wu, Ying Huang, Tengfei Li, Li Wang, Yongchang Chen, Weizhi Ji, Yu Zhang, Gang Li, Yuyu Niu. NeuroImage 247: 118799, February 2022, |
6. UNC-BCP 4D Infant Brain Volumetric Atlases
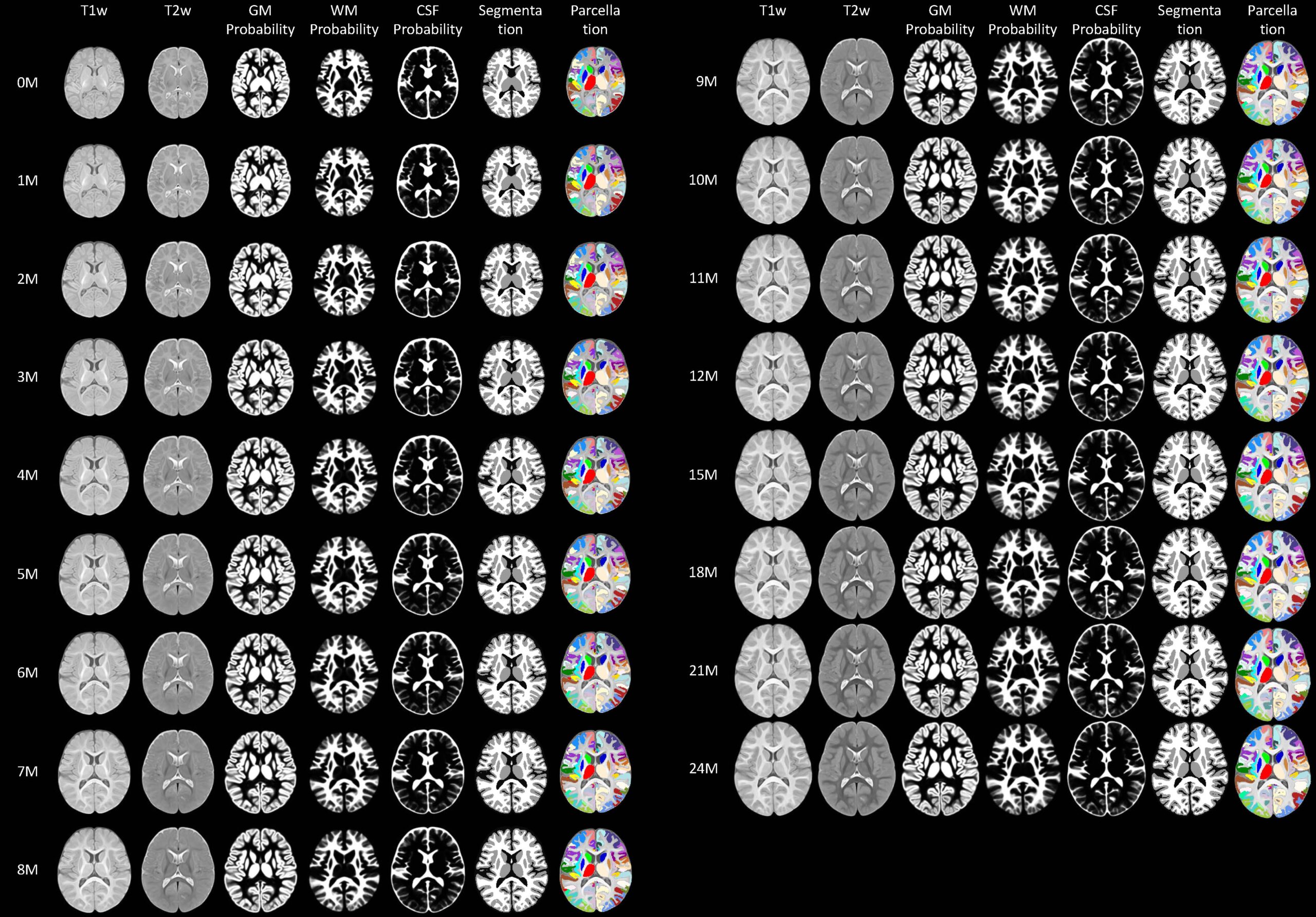 |
UNC-BCP 4D infant brain volumetric atlases include atlases at 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 15, 18, 21, 24 months, which include the following components: 1) group-representative T1w and T2w images, 2) tissue probability maps, 3) tissue segmentation maps, 4) cortical parcellation map, and 5) subcortical labels. These 4D atlases have a high spatial resolution, large age-range coverage (from birth to 2 years of age), and densely sampled time points. Specifically, 542 MRI scans with T1w and T2w sequences from 240 infants up to 26-month scan-age were utilized for our atlas construction. (Download on NITRC) A 4D Infant Brain Volumetric Atlas Based on the UNC/UMN Baby Connectome Project (BCP) Cohort. Liangjun Chen, Zhengwang Wu, Dan Hu, Ya Wang, Fenqiang Zhao, Tao Zhong, Weili Lin, Li Wang, Gang Li. NeuroImage 253: 119097, June 2022. |